For this project you will study a small molecule (hetero compound) that is complexed in a protein. You will determine if the hetero compound is in its minimum energy conformation when it is complexed. Additionally, you will describe the nature of the interactions between the small molecule and the protein that it is complexed with. To this end, you will
All the results will be assembled in an HTML document linked on your web site. Additional activities are included that review some of the topics covered during the semester. As the various parts of the assignment become due, you will post the results on your web site.
This assignment will help you learn the features of DSVisualizer, introduce you to some biomolecular chemistry databases, reinforce your previous work in Chem3D, and review protein structure and intermolecular interactions.
As you visit the various web sites, especially the PDB site, make sure you find out how they suggest citing their pages for the files you later incorporate in your project report. Also, see the CHEM 350 Manual on Citation Style.
See the Courseware page for the DSVisualizer download site and the Chemical Information list for database sources. The course Manual contains information on using DSVisualizer and Chem3D. Here is a brief review of protein structure.
1. Choose an organic heterocompound. In Spring 2009, each student will choose a drug molecule from the list below. All of these drugs have a protein target. As each hetero choice is made, I will put an X next to the name and then it will be unavailable to anyone else. Please remember to refresh your browser.
Go to one of the database sites (HIC-Up, PDB-At-A-Glance,RCSB PDB, Het-PDB Navi, or PDBSum) to find the 3-letter het code for your chosen molecule.
2. Next, choose a protein that complexes the heterocompound. You can find a protein in the sites mentioned above although it might be downloadable only at the main RCSB PDB site (use the heterocompound het code in a keyword search or use advanced search).
3. At RCSB PDB, click the Download pdb file icon button to the right of the checked file box and PDB code. Save the pdb file to your desired location.
Assignment:
Please
send me an e-mail with your choice of heterocompound. State the
name of the hetero compound and its het code; state the name and PDB
ID number of the protein. Do this before completing other parts of this assignment.
Be sure to inspect your protein and heterocompound file to be sure that the heterocompound is complexed correctly as explained below. If you have problems, please let me know ASAP.
3. Open the pdb file in DSVisualizer. See the Manual for adjusting to Ultra-high viewing quality in the Graphics window and other features of DSV.
Open a Hierarchy window.
Inspect the components of the molecular file by selecting each of the listed entities in the Hierarchy window and looking at the corresponding highlighted groups in the Graphics window. Expand each listing by clicking on the "+".
Hide or Delete the water molecules, if any, by left-clicking on "Water" in the Hierarchy list, right-clicking on the selection, and then from the pop-up menu choosing either to delete or hide (or simply uncheck the box).
If you have only one chain in your protein, it is probably called "A". If there is more than one protein in the quaternary structure, they will be listed as "B", "C", etc. Select each extra chain and delete it (Ctrl X).
The heterocompound(s) appear in different areas of the Hierarchy window: sometimes they are listed with a a letter (e.g. "A"); sometimes under the category "Chain"; other times it appears under the label "Heterocompound". There may be only one, or several, complexed molecules. You might have to search a bit for yours. Delete any duplicate heterocompounds so that only one remains in your file.
Ideally, your heterocompound is complexed somewhere within the protein 3D structure. If the heterocompound appears attached to the protein's periphery, as illustrated below, it may mean the compound is interacting with two protein chains. This complicates the analysis at the end (not greatly), so if you have another protein to choose, you should consider it.
Make sure the hetero compound contains all the atoms that are shown for its structure (except hydrogens).
| Amodiaquine | X | Pentostatin | |
| X | Bepridil | X |
Phenylbutazone |
| X | Diclofenac | X | Pyrimethamine |
| X | Diflunisal | X | S-Naproxen |
| X | Diphenhydramine | X | Tadalafil |
| X | Donepezil | Trifluoperazine | |
| X | Fluconazole | X | Trimethoprim |
| X | Gabapentin | X | Trimetrexate |
| X | Ketoconazole | X | Warfarin |
| X | Penciclovir |
1. Select the hetero compound ID code in the DSV Hierarchy window (notice that when you highlight the code in the Hierarchy window the corresponding molecule is also highlighted in the model window), copy it, and paste it in a new DSV model window. You have now "extracted" the hetero compound.
Carefully inspect the structure of the extracted compound and make sure it matches the known structure, with respect to all atoms except hydrogen, as found in some other source (such as the PDB site, Dictionary of Organic Compounds or a textbook). In some cases the heterocompound is "broken", in other cases it might be covalently bonded to one of the amino acid side chains-- please inspect it carefully.
2. A hetero molecule that contains aromatic sp2 carbons, and sp2 carbons generally, is sometimes mis-defined when imported into DSV. When hydrogens are added in DSV, a benzene ring can become a cyclohexane and C=O groups can become alcohols!
Also, when acidic or basic compounds are extracted, it is unclear whether or not the compound is charged. Generally, carboxylic acids are most likely in the form of a carboxylate and thus negatively charge under pysiological conditionsd. Likewise, amino-containing compounds are most probably positively charged. Phosphates will be negatively charged. These also may need to be fixed in DSV. Amino acids and peptides are zwitterions.
See this document (fix-geom.pdf) for instructions on fixing the hybridization and charge.
After making any necessary hybridization/charge adjustments, choose Tools | Hydrogens | Add/Show from the menu bar. Re-inspect the molecule for the correct hybridization, geometry and number of hydrogen atoms and charge. Save the molecule under a new file name in the MDL .mol file format.
Rotate and position the molecule in the window as a ball and stick model. This file is your "extracted" hetero compound. Using a viewer window of a size that will fit eventually into a Word printout, save the molecule in the .msv format. Then save the molecule in the jpg format. (Change the background color of your graphics window to white or a light pastel color so you don't waste printer ink when you print your final document.) Use high graphics settings for the jpg images (do not use "screen shots"). See the Manual for more detailed instructions on saving images from DSV.
For all image files you save for later incorporation into the final presentation document, the size should be large enough for details to be viewed. The image should nearly fill the space in the window. For the image above, the size should be about half the width of a Word document page. The image size is chosen while saving in DSV. The image is better quality if the Graphics window is sized properly before the file is saved. Your image will be judged by its quality.
Draw a Lewis structure of your hetero compound. Include stereobonds if known; include all non-bonded electrons. The drawing should look professionally drawn and should be of a size comparable to the ball and stick rendering from DSV, above. Remember, if you have a carboxylic acid it most likely exists as the carboxylate at physiological pH and so you must draw it in that form. Save the drawing in the gif format.
In the Figure below, the amino acid residues in the protein are shown near the top of the expanded Hierarchy window listing. The heterocompound is listed separately under "A" as ADP918.
Duplicate this view of the space-fillling hetero molecule in its protein binding site (make sure no part of the structure is selected (that is, colored in yellow highlighter) with the heterocompound prominently displayed):
1) In the Graphics window, choose View | Display Style. Set Atom | None and Protein | Solid Ribbon and Color | Secondary Type.
2) In the Hierarchy window, select the hetero compound; in View | Display Style set Atom | CPK. While the molecule is still selected, choose Tools | Hydrogens | Hide.
3) Make sure that you have deselected the heterocompound and it is shown in its proper CPK colors. Save the file in the msv format.
4) Again save the file: choose File | Save As.... | Image files and then save as a jpg file. The image should be large enough to nearly span the width of a Word page.
Begin a Word document in which you describe what you have done so far and what programs you have used. Be liberal in your use of words! The heterocompound should be given both its chemical name and het code. The protein should be identified by name and PDB code. Include a document title. Name the file proteinXXX.doc, where XXX are your initials. Incorporate the images described above that illustrate your explanation. Images should have informative captions. For images that do not occupy the entire width of the page, text-wrap around the image. Remember to properly cite the website that you obtained your protein file from. All the protein/heterocompound sites tell you how to cite them. Construct a new web page titled Protein-Modeling within a new directory Protein that is within your Projects directory . Link to Protein-Modeling from your CHEM 350 Projects page. All files for this Assignment will go into the Protein directory. Insert the ball-and-stick image file of your extracted hetero compound on the Protein-Modeling page and provide its name as a caption underneath the image. Do the same for the Lewis structure of your hetero compound and for the protein-hetero complex. Hyperlink your Word document on the page, using informative text. Insert the image file of your protein-hetero complex on your Protein-Modeling page. Provide an informative caption. [Remember to properly cite the website that you have obtained the protein-hetero file from, if you have not already done so. All the protein/heterocompound sites tell you how to cite them.] Update your Word document and link it to your Protein-Modeling page. |
Open the extracted hetero compound .mol file in Chem3D. Save it in the default Chem3D file format.
If you have an aromatic ring, check the bond order: it should be 1.5. If it is not, it may have reverted to a cyclohexane.
Perform a single-point energy calculation on the extracted hetero compound. DO NOT minimize the energy/optimize the geometry.
Copy the "out" message text for the steric energy. Before pasting the text into your on-going proteinXXX.doc Word document, embed an Excel worksheet into Word. Then paste the text into cell A1 of the spreadsheet that is now embedded in Word. You can then proceed to convert the text to columns as necessary. Be sure your table title and column headings are informative. Make sure the worksheet fits within the margins of the Word page. Space the columns so the text is not compressed in a small space and use text-wrapping in columns as necessary. This should look professionally done.
Copy (not interactive) the hetero compound from Chem3D and paste it in the Word document. The width of the image should be sufficient to see some detail but not overly large. Left justify the image. Provide an informative caption.
This value is the steric energy of the compound when it is complexed within the protein. It is probably not a local or global energy minimum as found for the same compound in the gas phase where there are no intermolecular interactions.
Save the Chem3D file again, with a new name, before minimizing its energy (a geometry optimization). Perform an MM2 energy minimization. This is your "energy-minimized" hetero compound. Copy the steric energy information and paste it into the embedded worksheet in Word. The before and after steric energy information should be in two parallel columns; there should be only one column of labels (Stretch, Bend, etc.).
Copy (not interactive) the minimized structure from Chem3D and paste it in the Word document next to or below the "before" image. The two images should be the same size. Provide an informative caption.
This energy calculation corresponds to an energy minimum for the compound in the gas phase.
In several paragraphs, explain the purpose of the before/after comparisons and then expicitly compare the steric energy components and values before and after geometry optimization. Discuss each steric energy term separately, making sure you define the term. Which term contributes the most/least to the total steric energy? Try to account for the component steric energy change(s) with specific reference to your molecule.
Save the Chem3D and Word files.
On the web page titled Protein-Modeling, upload the up-dated, spell-checked Word .doc file. [There should be only one Word document that you continually add to.] Make a link to the latest Chem3D file. Note: When you click on the hyperlink to your Chem3D file, you will see only text. This is because the web browser isn't Chem3D and so doesn't open the display. It just shows the text behind the display. It's fine. If anyone wanted to see your display, they would have to save the file first and then open it on a PC with Chem3D. |
1. In Chem3D, open the files containing your extracted hetero compound and your energy-minimized hetero compound. If Chem3D added Lone Pairs to your compound, hide them. Choose either of the two files and save it under a new Chem3D file name.
Copy and paste the other structure into the newly named file. Both molecules will be in approximately the same orientation. You can rotate one of them by selecting it and press the SHIFT key while rotating (or translating). Unselect and rotate both compounds so that you get a clear view of most of the atoms.
2. Choose View | Model Explorer, which opens up a panel with Fragments listed. The Fragments are the two molecules in the window. You can select one of the Fragments in the Model Explorer and the corresponding atoms are selected in the model (just like DSV). To move or rotate the selected fragment only, hold down the Shift key.If there are several fragments listed, they might be lone pairs, even if they are hidden. Check by expanding the Fragment box. Ignore the extra list of Fragments or delete them.
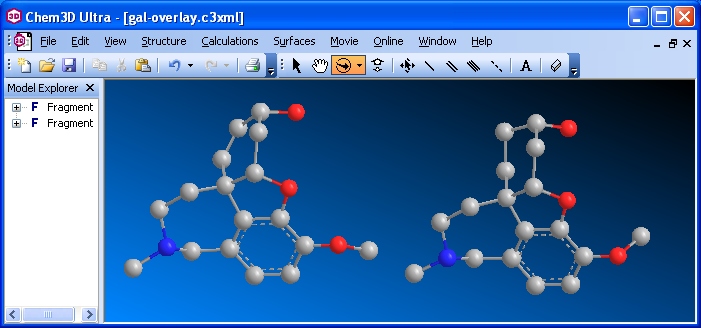
Extracted and minimzed hetero compounds before Overlay (Ultra/ version 10)
Select one of the fragments. From the menu bar choose Structure | Overlay | Set Target Fragment. Select the other Fragment. From the menu bar choose Structure | Overlay | Fast Overlay. Hide the serial numbers by unchecking Show Atom Map Labels.
Remember, one of these structures corresponds to the compound's conformation while complexed within the protein; the other structure corresponds to a gas phase energy minimum. They most probably will not, nor be expected to, overlay exactly.
3. Rotate the two overlaid molecules simultaneously so a visual comparison can be made easily. In most cases the hydrogens will not overlay. If you only have a few hydrogens in your compound, you should show them. If you have many hydrogens, you should choose to show Polar Hydrogens only. Keep track of which model is the extracted original and which is the energy-minimized.
Rotate the overlaid structures so you can see which atoms deviate the most. In the example shown below, the CH3 on the O attached to the aromatic ring does not coincide in the two structures (it almost seems as though there are two CH3 groups bonded to the O). Notice also the OH in the upper right -- they do not coincide either. You would see some other minor deviations as you rotate your model.
Save the file as "overlay" in the default Chem3D file format. From Chem3D save the file as "overlay" in the jpg graphic format.
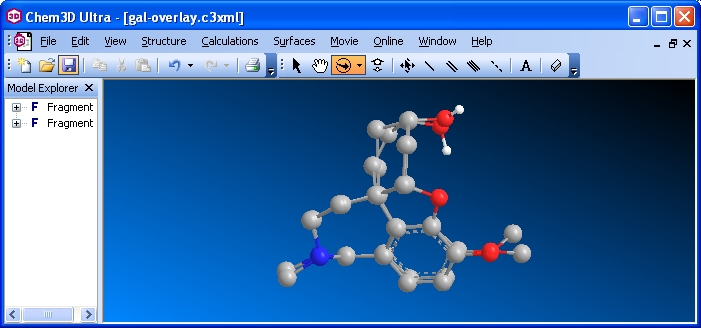
Images of the overlaid extracted and minimzed hetero compounds
Insert the overlay graphic file in your Word document. Write a paragraph (consisting of many sentences) describing what you did. Then comment on the "goodness of fit" of the extracted and minimized molecular conformations. Comment specifically on the structural parts of your two molecules that do and do not overlay. Update the linked Word file on your web page. Submit a printout of the Word document. Zip all the files that constitute the Protein-modeling web page. Submit it to WebCT. |
1. Go to the PDSum site and enter the PDB code for your protein in the search box. In the first results page click the [Protein] tab and look at the wiring diagram for the primary/secondary structure of the protein chain that contains your hetero compound. (Make sure that you are looking at the correct chain if you have a multi-chain protein.) The red dots, if any, point to the amino acid residues that interact with the ligand. The red rectangles (or colored triangles), if any, indicate the amino acid residues at the protein's catalytic active site. The blue dots, if any, are residue interactions.
Save the gif file of the diagram. (If you want to print the graphic, the quality is better when the graphic file is viewed by itself in a new browser window or if you open the accompanying Adobe pdf file.)
Click the [Ligands] tab to view a "LIGPLOT" of ligand (heterocompound) interactions with the protein. There may be many ligands listed and there may be duplicates of your heterocompound especially if your original protein file had a quaternary structure -- be sure you select the correct one.
Click on the Adobe pdf hyperlink to see a better quality image of the ligplot. The amino acid residues shown interacting with the heterocompound in this ligplot image should match the residue symbols in the wiring diagram described above. Take a "snapshot" of the diagram in the pdf file using the Adobe snapshot tool. Save it in a graphics program as a jpg file.
2. Open your protein/heterocompound pdb file in DSV. As you complete each of the following items to your satisfaction, save the file in the msv format which will preserve your viewing options and molecular orientation, unlike the mol or pdb formats that open in the default line display style.You should probably save several sequential versions in case you need to make changes later.
For all image files you save for later incorporation into the final presentation document, the size should be approximately 800 pixels wide and the molecule image should fill the space (i.e. it should be large enough to see the detail). The image size is chosen while saving in DSV. The image is better quality if the Graphics window is sized properly before the file is saved. Please use a light-colored background.
Display the Protein as a Solid Ribbon and color by "Molecule". In the Atom tab, turn the display to "None". Your protein should be a pale gray against a light background. Save.
Open a Hierarchy Window. Select your hetero group from the hierarchy window and choose to display the atoms in a ball-and-stick style in the elements' colors. (You can't see the selection in the 3D window until you choose an atom display style.) This is similar to the view in Part I.
In the Hierarchy window, expand the protein chain to see the list of amino acid residues. Select by residue number each of the amino acid residues that are specified in the complexing site (and catalytic site, if applicable) as found in the PDBSum wiring diagram. Display them in stick style in a pale yellow color. While the residues are still selected, right-click the mouse button and choose Label... . Choose Object: Amino acid, Attribute: Name, and Font: Courier 8. [It is easiest to choose all the relevant amino acids first, then apply the color and labels to them all in one operation.] Choose a dark color for the name labels.
Turn off the protein display. Rotate the window contents (only the hetero compound and amino acid residues should be visible) so that the view matches as closely as possible the LIGPLOT diagram. This can be quite difficult since the ligplot is highly stylized and is trying to present 3D information in 2D. The residues that are denoted by "eyelashes" are usually above or behind the plane of the ligplot. Remember, to rotate around the Z axis in DSV hold down the [Shift] key.
Be sure to note any discrepancies between the residues specified in the wiring diagram and the LigPlot.
Show all hydrogens in your heterocompound.** Choose Stucture | Monitor | HBond. You should see dotted green lines appear between atoms that are hydrogen-bonded. Ideally, you should see hydrogen-bonds from the same residues to the hetero compound that are indicated in the ligplot. The default distance monitor for hydrogen-bonds in DSVisualizer is 2.5 A. You can increase the distance by selecting HBond Monitor in the Hierarchy window, right-clicking and selecting "Attributes of HBond Monitor...". Change the distance to include the largest hydrogen-bonding distance to the hetero compound as shown in the LIGPLOT. (You can also change the hydrogen-bond to a darker color for better visibility.) Try to rotate the image to show these hydrogen bonds and the other atoms as they appear in the LIGPLOT.
Save the view as an msv file and jpg.
** If the hydrogen-bonds don't show as in the ligplot, it may be that DSV added the H's in a "wrong" position. Remove the hydrogens to see if the H-bonds now display properly.
Set the protein display to Solid Ribbon, Color by Molecule and choose a medium gray for the protein on a light color background. Make sure all the elements of the protein-hetero complex display to best advantage. Save the view as an msv and a jpg file.
This is an example of what the images should look like (approximately).
Incorporate the images and the explanations of what you have done into your Word document. Be sure to explain what is represented in the Wiring Diagram and LigPlot. Tell what you have done in DSV to correspond with those. Do not assume your reader has ever seen these before. The Word document is already linked to your web page -- make sure you upload the revised document to your web site. Make all necessary revisions to the Word document to improve it from your previous submissions. |
In the previous part, you identified the amino acid residues in the protein that interact with the heterocompound. In this part, you will specify the interacting groups and the type of interaction. Here is a document showing the amino acid residues and the type of interactions.
Construct a table in your Word document, continued from a preceding part. In the first column, list the amino acid residues in their primary structure order, and their position numbers, that interact as shown in the LigPlot (ideally the Ligplot and Wiring diagram should be identical; in practice they might not be). In the second column, specify the atoms in the heterocompound that interact with the residue. In the third column, state the interaction (see the Amino Acid and Protein document discussed in class and in the Manual). For the protein, be sure to differentiate between an interaction with the amino acid residue side chain or with the backbone peptide bond components (C=O and N-H).
It might be difficult to identify the interactions between the heterocompound and the "eyelash" residues in LigPlot. Generally, these are hydrophobic interactions, unless the residue is clearly an electrostatic or hydrogen-bonding residue. Inspecting your three-dimensional DSV model will help you decide the interactions.
Here is an example. It is an extension of the example linked in the preceding part.
Update the Word document and upload it to your website. |
There are three important citations that belong in your Bibliography section: one is to the PDB site where you downloaded your protein-heterocompound pdb file, another is to the journal article that first reported its structure, and the third is where you obtained your wiring diagram and LigPlot. All citations should be complete and in the style shown in the Manual. Use reference numbers enclosed within brackets in the text that refer to the citations in your Bibliography section. Any other information that you incorporate should also be properly acknowledged and cited.
1. Nearly all the biochemical molecules sites give some instruction on how to cite their site. For the RCSB PDB site, look for the box that says "In citing the PDB...." and use the style given there.
2. At the RCSB PDB site, perform a keyword search on your hetero compound. Click on the protein icon corresponding to your protein-hetero complex. Acess the Structure Summary page. Part of the information on this page concerns the Primary Citation for the original publication of the protein-hetero complex. There are two links to Abstracts. Choose the one that will allow you to "View PubMed Abstract at NCBI".
On the resulting page there should be the abstract. Near the top there should be at least one link to the journal**. You might have to do some exploring to finally access the full-text article (preferably as a .pdf file).
In your Word document Bibliograpy section, cite the original journal article. Follow the guidelines in Chem350 Citation Style guide. Be sure the hyperlink in Word is an active link. Access the journal article and save it.
**If there is no journal link or if the work has not yet been published, choose a different protein that complexes your heterocompound. You can still cite the original article for your combination (but no hyperlink) and also cite and hyperlink this secondary article. Of the proteins that complex your heterocompound, at least one should be suitable. Please let me know.
3. Remember to cite the PDBsum site.
Update the Word document and upload it to your website. |
By now you should have a complete Word document of this assignment. Make sure all elements of the assignment (Parts I-II) are present, properly displayed, and in the order they were assigned. Complete any parts that you left unfinished. Be sure that the introductory paragraph includes the protein's name and PDB ID code and the hetero group's name and HET abbreviation. Include some information on the occurrence and function of the protein in nature. What is the hetero compound's function in this protein (more than one sentence is required -- it should appear as though you have actually researched this)?
Include explanatory section headings as given in the assignment.
Construct paragraphs explaining what you have done in each Part and that tie the various sections and images together in the document. You should be telling a complete story, from beginning to end, about what you did for this project. Do Not repeat the instructions and Do Not give details such as which menu commands you chose, keys pressed, etc. You will be graded also on your final submission, so if you have made errors earlier, they should be corrected.
Formatting
Save the Word file as an .htm file. Be sure to include the <title>. Upload the htm and the newly-created subdirectory of subsidiary files to your web site. Link it from your Protein Modeling page. Check that the page displays properly in both Explorer and Netscape/Firefox. Please differentiate the links to the the Word .doc document and the Word .htm document so that it is obvious what is linked.
Upload your revised Word .doc document. It should still be linked to your Protein Modeling page.
Print the entire Word .doc document and submit to my Chemistry Department mailbox (or underneath my office door) by the due deadline.
Submit to WebCT a zip file containing all files saved for this project.
| Chemistry 350 | George
Mason University |